-
My cart's content
-
Billing address
-
Shipping address
-
Payment method
-
Confirm order
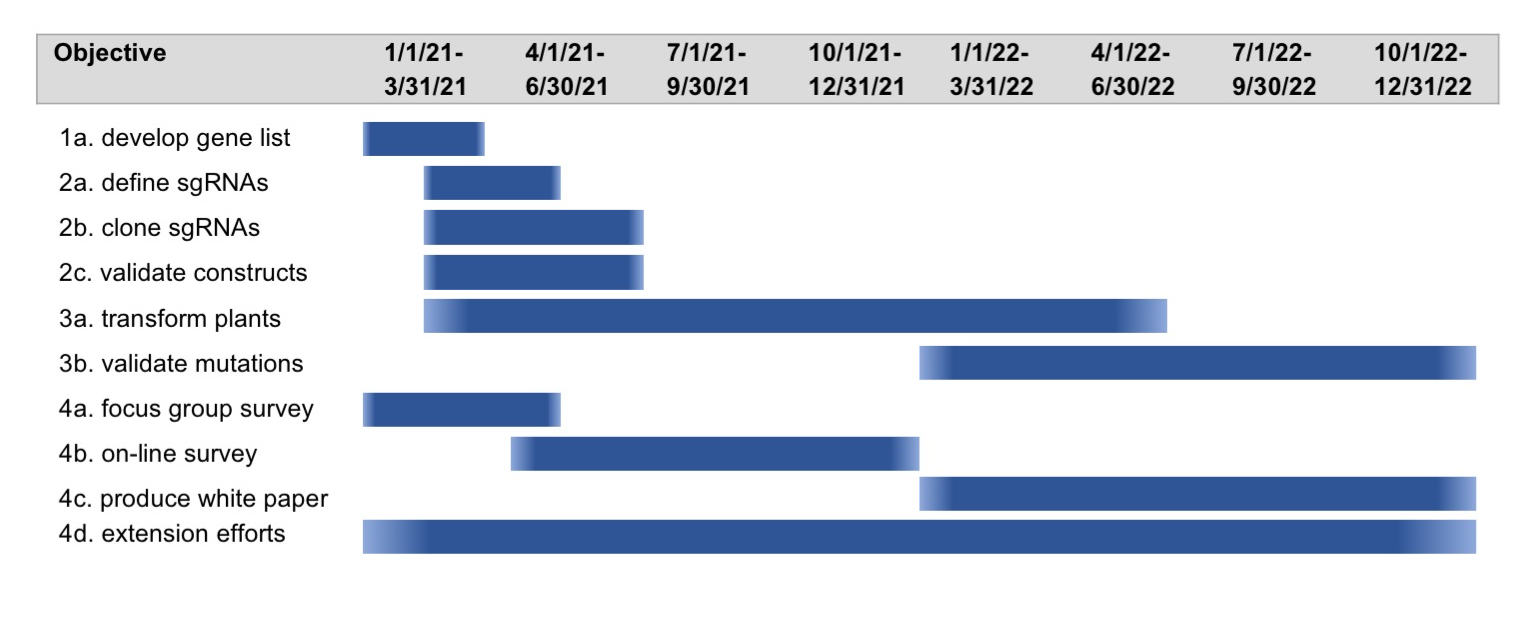
The U.S. citrus industry is facing unprecedented challenges from the spread of citrus greening disease (i.e Huanglongbing or HLB) in citrus-growing states like Florida, Texas and California. While traditional breeding approaches using natural variation are one way of introducing HLB tolerance into select cultivars, alternative approaches to rapidly generate new cultivars that are resistant/tolerant to the HLB-causing bacterial pathogen would be extremely valuable. The overall goal of this two year project is to utilize the extensive transcriptomic and proteomic information available for HLB infected citrus to develop a large-scale population of CRISPR/Cas9 gene-edited Valencia plants that can be screened for tolerance to HLB. This project will provide a sustainable resource (i.e. collection of citrus mutants) for the citrus research community that can also be screened for resistance to other diseases and for other value-added traits in the future, and thus accelerate discoveries that would benefit the U.S. citrus industry many years after the funding period is completed. As part of this project, we also propose to study the economic and societal impact of using gene-editing technologies like CRISPR/Cas9 to create new citrus cultivars, with a focus on increasing acceptance of gene-edited crops by consumers and growers. In the longer term, the mutations identified as enhancing defense against HLB can then be engineered into other commercially important citrus cultivars and field-tested in Florida and California.
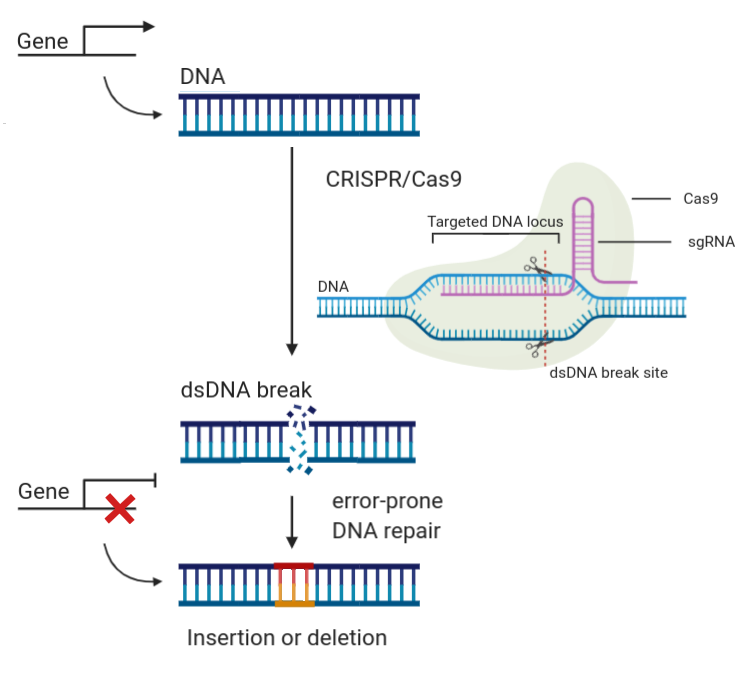
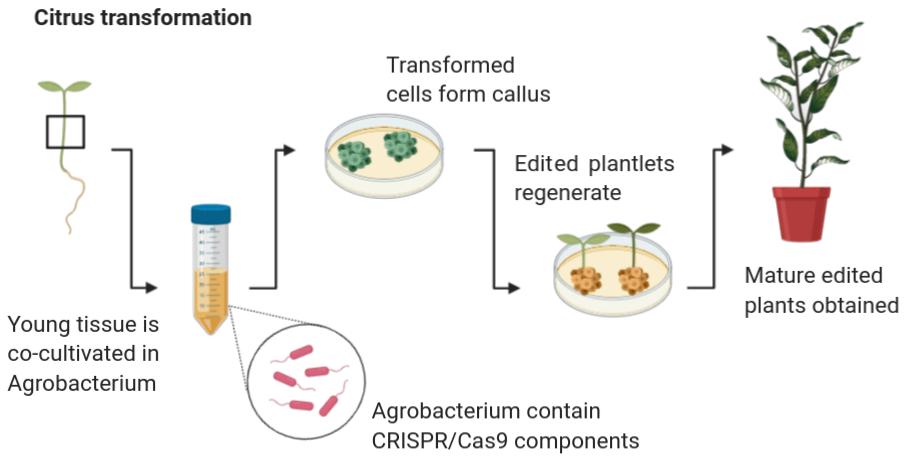
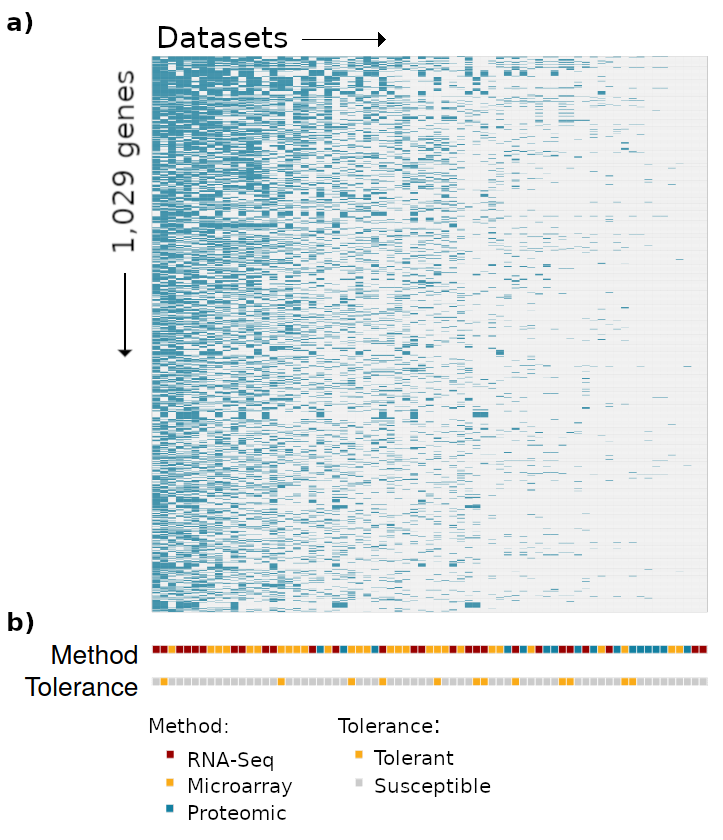
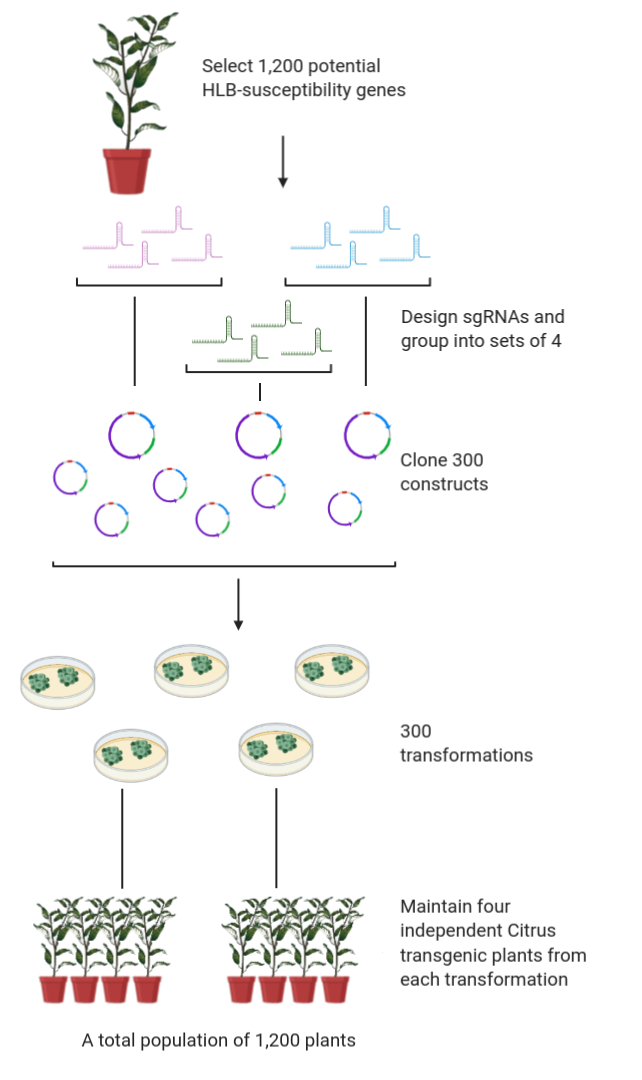

Over 25 years of research experience in plant developmental and molecular genetics.

Experience in optimizing novel genome-engineering technologies in plants, with a focus on Citrus.

Expertise in consumer acceptance of biotech foods, demand and consumer perception of fruits and vegetables.

